Fiber Tracking
Fiber tracking is a technique used in diffusion MRI to reconstruct the pathways of nerve fibers within the brain. It is based on the principle that water molecules tend to diffuse more readily along the axons of nerve fibers than across them. By measuring the diffusion of water molecules using diffusion MRI and applying advanced computational techniques, it is possible to create 3D reconstructions of the pathways of nerve fibers within the brain, known as “tracts.”
Before practicum on Friday, please complete following:
- Watch a video about tractography
During practicum on Friday:
Review of last weeks materials (5 min)
- Commissural pathway (X-axis): pathways connecting left-right cortical regions
- Association pathway (Y-axis): pathways connecting cortical regions within the same hemisphere
- dorsal: AF, SLF, FAT
- ventral: IFOF, UF, ILF, MdLF, VOF
- medial: cingulum
- Projection pathway (Z-axis) pathways connecting cortical regions to basal ganglia (caudate, putamen, globus pallidus), thalamus, brainstem nuclei
Introduction to tractography (15 min)
- First tractography by Basser ISMRM 1998, Poupon et al MICCAI 1998, Mori et al Ann Neuro 1999, Conturo et al PNAS 1999.
- Fiber tracking method
- Input: (1) local fiber orientation(s), a.k.a. fixel (2) termination metrics, e.g., anisotropy
- Output: sequences of 3D coordinates
- Deterministic vs probablistic

Local fiber orientations derived from dMRI
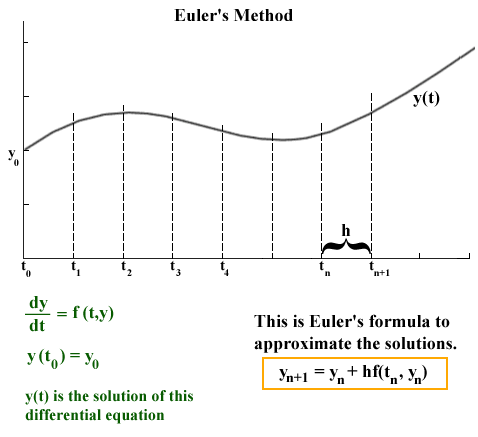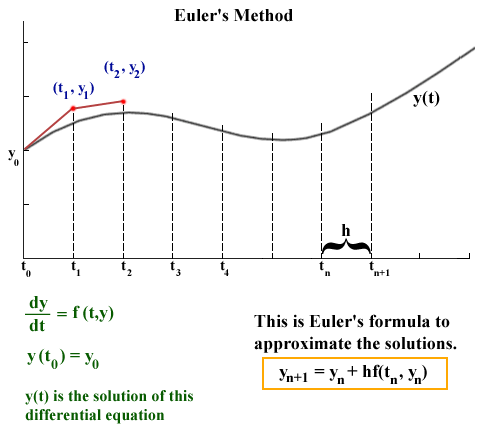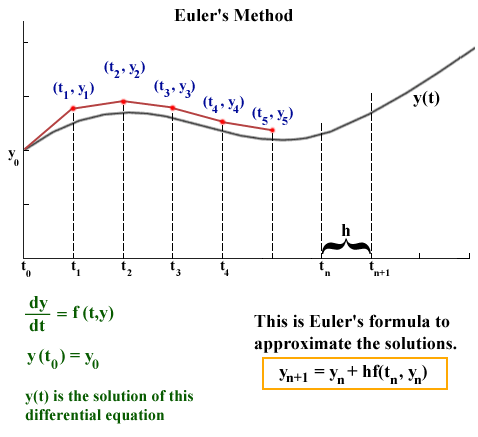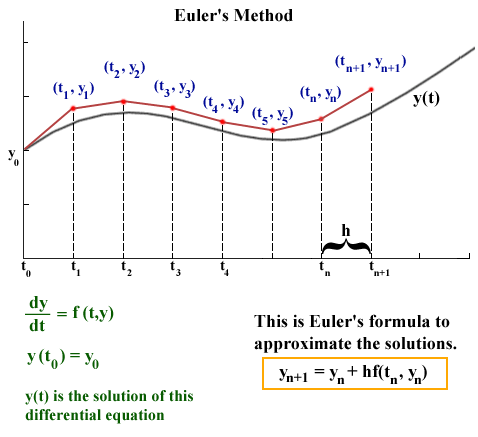
source: http://jmahaffy.sdsu.edu/courses/f00/math122/lectures/num_method_diff_equations/nummethod_diffeq.html
- Hands-on:
- 100206.src.gz.gqi.1.7.fib.gz
- fiber tracking using GUI
- anisotropy threshold
- angular threshold
- step size
- minimum and maximum length
- fiber tracking using CLI
dsi_studio --action=trk --source=*.fib.gz --parameter_id=c9A99193Fb803FdbF041b96438813cb01cbaCDCC4C3Ec > log.txt
Errors in tractography (15 min)
- What is good/bad tractography?


reference: Fernandez-Miranda JC,Neurosurgery. 2012 Aug 1;71(2):430-53.
- Error near the brain surface
- Mostly due to DWI processing and modeling
- eddy current or susceptibility distortion
- diffusion models
- Solutions: (1) better acquisition (2) better modeling
- Mostly due to DWI processing and modeling
- Error in deeper white matter
- Mostly due to limitations in fiber tracking algorithms
- false routing (a.k.a crossing/branching problem)
- gyral bias
- Solutions: (1) better spatial resolution (2) atlas-guided false fiber eliminations
- Mostly due to limitations in fiber tracking algorithms

reference: Yeh, Fang-Cheng, et al. “Tractography methods and findings in brain tumors and traumatic brain injury.” NeuroImage 245 (2021): 118651.

reference: Wu, Ye, et al. “Mitigating gyral bias in cortical tractography via asymmetric fiber orientation distributions.” Medical image analysis 59 (2020): 101543.
Region-based fiber tracking (15 min)
- Hands-on:
- Demonstrate Region types: seed, ROI, ROA, END, terminative
- ROI: a filtering region that filters IN tracks
- ROA: a filtering region that filters OUT tracks
- Seed: the starting location of fiber tracking
- END: a filtering region that filters IN tracks ending in the region
- Not-END: a filtering region that filters OUT tracks ending in the region
- Terminative: cut tracks that enters the region
- Recommended steps to find a pathway between left and right V1
- Assign HCP_MMP:L_V1 and HCP_MMP:L_V2 as ROIs
- Adding seed regions to speed up
- Adding negated-dilated NOA
- Adding ROA/END/NOT END to refine
- Fiber tracking using CLI
- Rename the regions files to include “mni” in the file name, and DSI Studio will load it as an MNI regions
2 ROIs
dsi_studio --action=trk --source=*.fib.gz --parameter_id=c9A99193Fb803FdbF041b96438813cb01cbaCDCC4C3Ec --roi=HCP-MMP:L_V1 --roi2=HCP-MMP:R_V1
2 ROIs + seed
dsi_studio --action=trk --source=*.fib.gz --parameter_id=c9A99193Fb803FdbF041b96438813cb01cbaCDCC4C3Ec --roi=HCP-MMP:L_V1 --roi2=HCP-MMP:R_V1 --seed=tract_mni.nii.gz
3 ROIs + seed + NOA
dsi_studio --action=trk --source=*.fib.gz --parameter_id=c9A99193Fb803FdbF041b96438813cb01cbaCDCC4C3Ec --roi=HCP-MMP:L_V1 --roi2=HCP-MMP:R_V1 --seed=tract_mni.nii.gz --roa=tract_mni.nii.gz,dilation,dilation,dilation,dilation,dilation,dilation,negate
- Examples (Optional)
- Region Manual(source: Schneider lab)
- TractEM
Assignment:
1. Combine automatic fiber tracking and region-based fiber tracking
- Use automatic fiber tracking to map left corticospinal tracts on 100206.src.gz.gqi.1.7.fib.gz
- Identify false results and use MNI-space ROI or ROA regions to improve the tractogram.
- Use command line to repeat the same on subject-space FIB file(Download 3~5 FIB files)
2. Mapping difficult pathways using region-based fiber tracking

- Download HCP1065 1-mm FIB file
- Use region-based fiber tracking, map the orbital connections in nuclei 5.
- Use region-based fiber tracking, map the temporal connections in nuclei 5.
- Use region-based fiber tracking, map the occipital connections in nuclei 5.